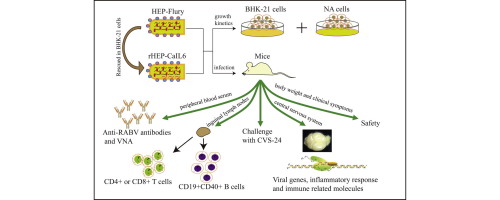
Abstract Rabies virus Recombinant, negative-strand RNA viruses, infect neurons via axon terminals and spread transsynaptically in a retrograde direction between neurons. Rabies viruses whose glycoprotein (G) gene is deleted from
Cusabio Other Organism RecombinantsCusabio Other Organism Recombinants
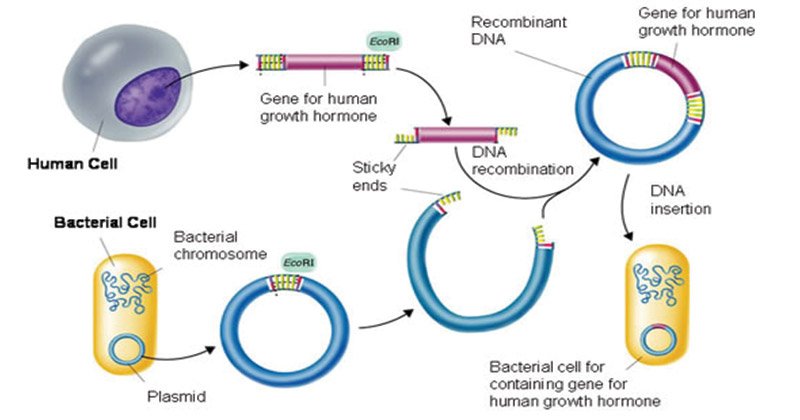
An organism refers to a living being that has an organized structure, can react to stimuli, reproduce, grow, adapt, and maintain homeostasis. An organism would therefore be any animal, plant,
Cusabio Lassa virus RecombinantCusabio Lassa virus Recombinant
Introduction Lassa virus, which is a member of the Arenaviridae family of viruses, causes acute viral hemorrhagic fever. It was first reported in 1969 in the town of Lassa, Borno
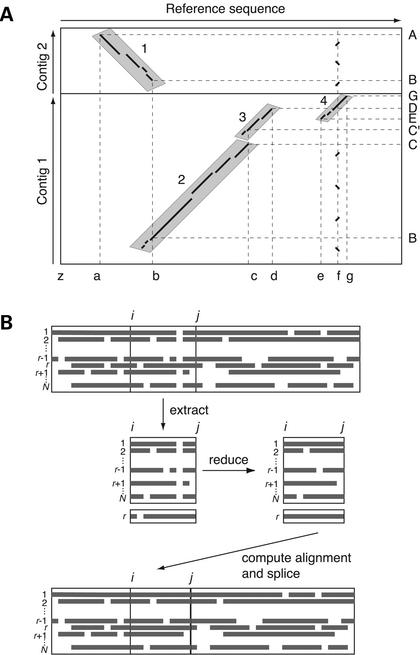
MultiPipMaker and supporting tools: Alignments and analysis of multiple genomic DNA sequences.MultiPipMaker and supporting tools: Alignments and analysis of multiple genomic DNA sequences.
Analysis of multiple sequence alignments can generate vital, testable hypotheses in regards to the phylogenetic historical past and mobile perform of genomic sequences. We describe the MultiPipMaker server, which aligns
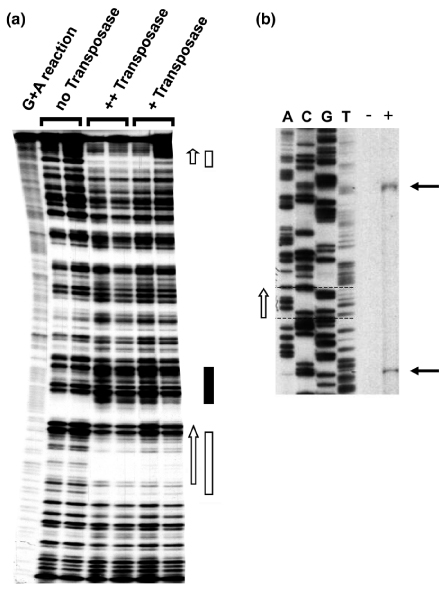
The DNA transposon Minos as a tool for transgenesis and functional genomic analysis in vertebrates and invertebrates.The DNA transposon Minos as a tool for transgenesis and functional genomic analysis in vertebrates and invertebrates.
Transposons are highly effective instruments for conducting genetic manipulation and functional research in organisms which are of scientific, financial, or medical curiosity. Minos, a member of the Tc1/mariner household of
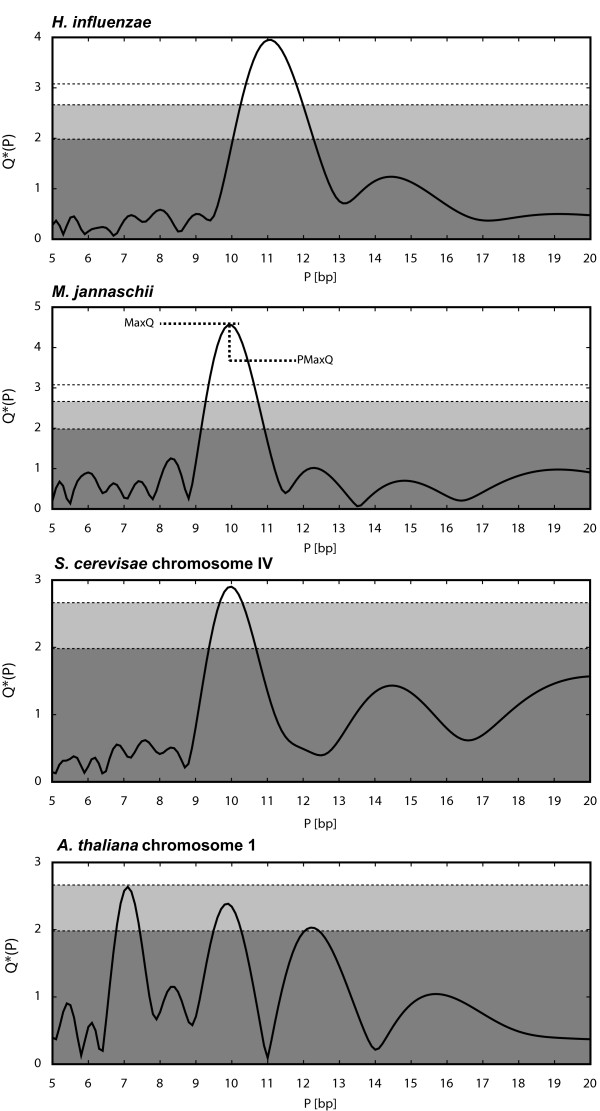
PerPlot & PerScan: tools for analysis of DNA curvature-related periodicity in genomic nucleotide sequences.PerPlot & PerScan: tools for analysis of DNA curvature-related periodicity in genomic nucleotide sequences.
BACKGROUNDPeriodic spacing of brief adenine or thymine runs phased with DNA helical interval of ~10.5 bp is related to intrinsic DNA curvature and deformability, which play essential roles in DNA-protein
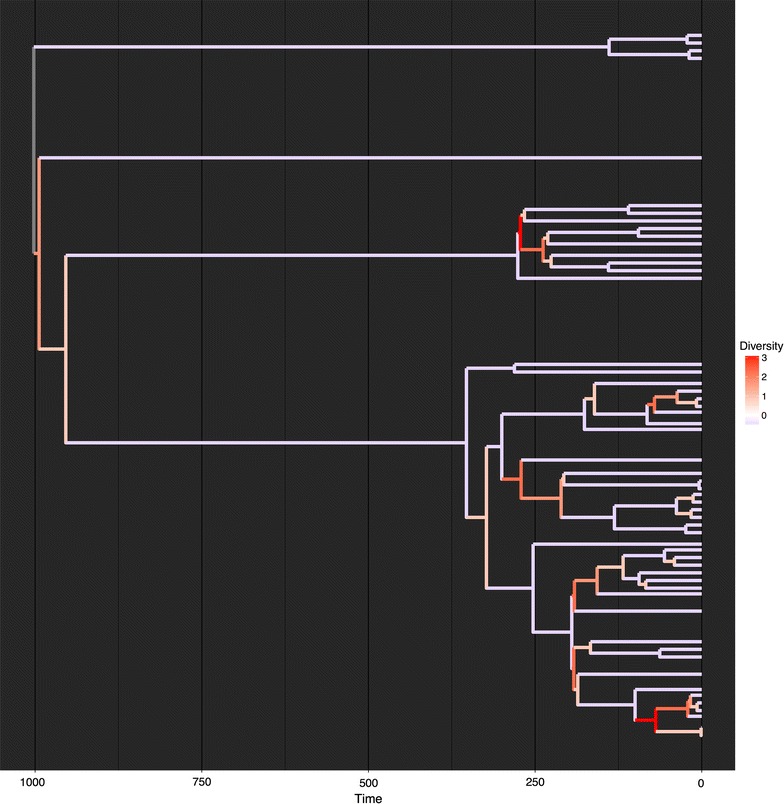
Tree simulation is an important tool for exploring the processesTree simulation is an important tool for exploring the processes
TreeMan is an S4 class that encodes a phylogenetic tree utilizing a node listing. The benefit of a node listing is the sooner computational processing, and the prepared capability to
Tree simulation: generating trees using different models of evolutionTree simulation: generating trees using different models of evolution
Tree simulation is a crucial instrument for exploring the processes that will have generated biodiversity. A typical instrument for simulating trees is the start–dying simulation or equals-rates Markov mannequin (ERMM)
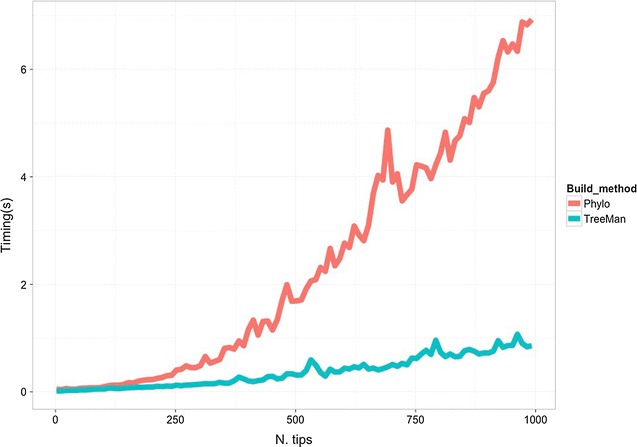
The TreeMan object in R is an S4 formal classThe TreeMan object in R is an S4 formal class
The TreeMan object in R is an S4 formal class whose foremost information slot is an inventory—which in R is a vector whose components might be named. All nodes in
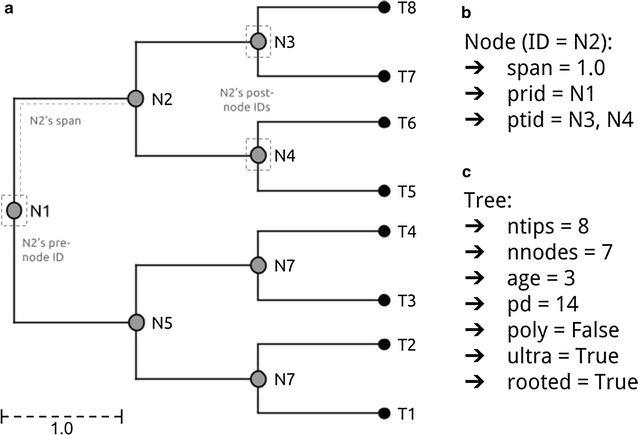
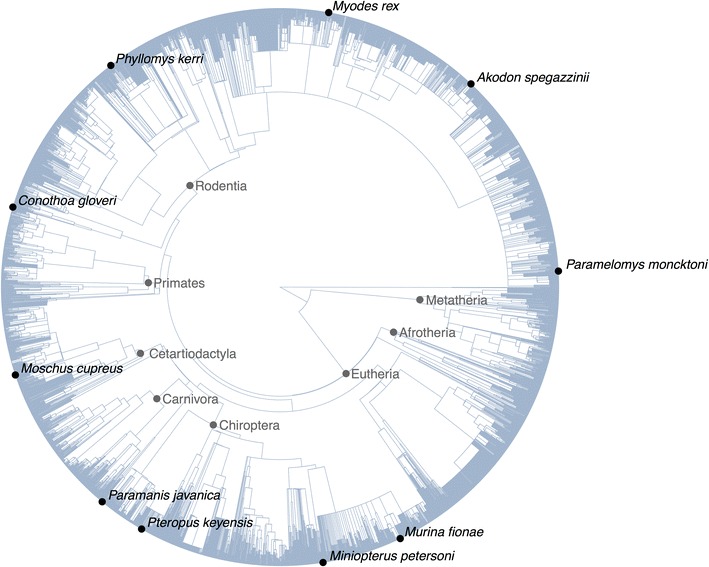
treeman: an R package for efficient and intuitive manipulation of phylogenetic treestreeman: an R package for efficient and intuitive manipulation of phylogenetic trees
BACKGROUNDPhylogenetic trees are hierarchical constructions used for representing the inter-relationships between organic entities. They are the commonest device for representing evolution and are important to a variety of fields throughout